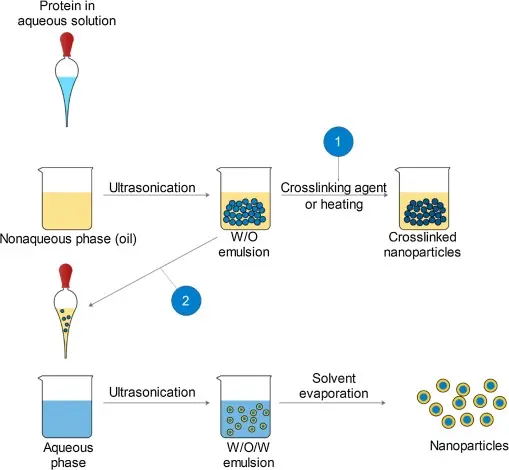
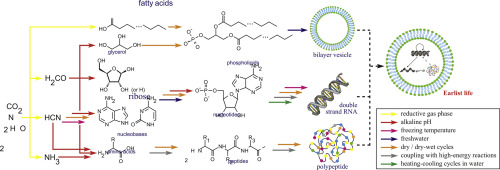
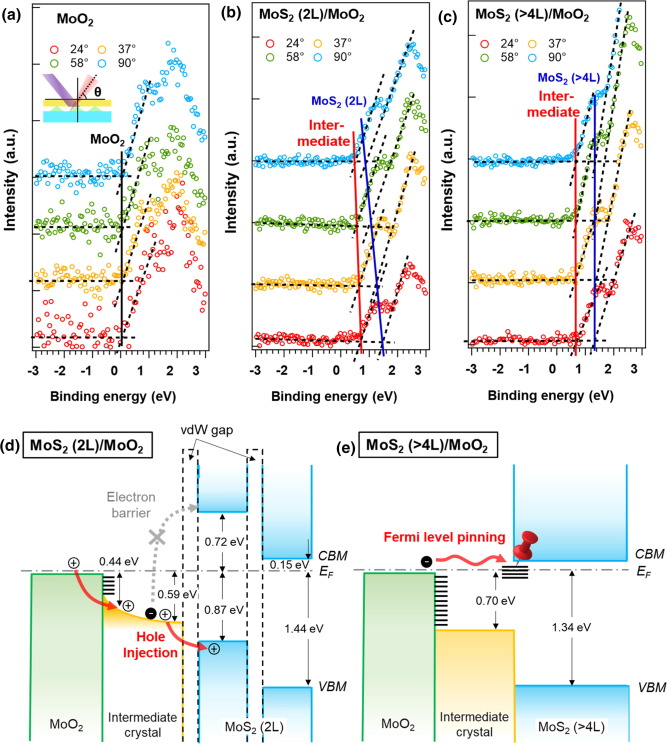
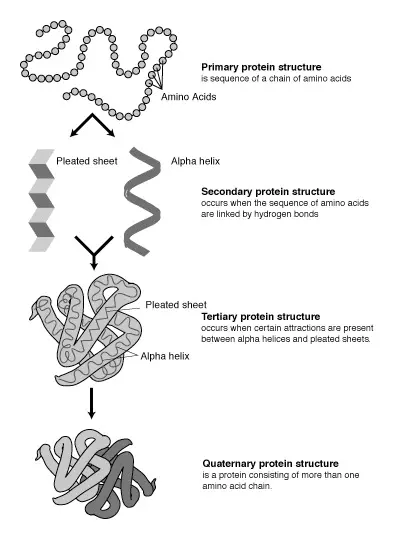
Getting Started with Insulin Crystallization
Crystallizing insulin is a straightforward yet highly technical process used in pharmaceutical applications and structural studies. The first step involves preparing an insulin solution by dissolving the hormone in an appropriate solvent, usually water or saline. This solution is then carefully adjusted for temperature, pH, and concentration to ensure optimal crystallization conditions.
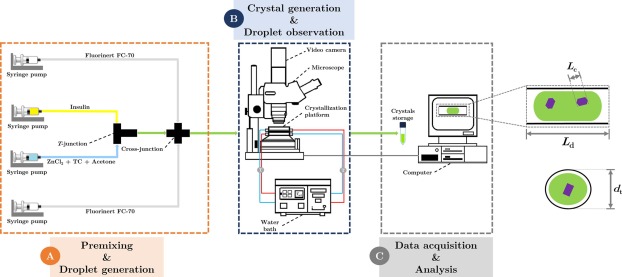
Once the solution is ready, crystallization is induced by controlling factors like concentration, temperature, and the addition of precipitating agents, such as salts. These changes cause insulin molecules to self-assemble into regular and ordered crystal structures. This process may take time, but careful monitoring ensures the creation of high-quality crystals.
After the crystals form, they are characterized using techniques like X-ray diffraction to determine the three-dimensional structure of insulin. X-ray diffraction provides detailed information about the arrangement of atoms and the formation of disulfide bonds in the insulin molecules.